T H E O R Y
Introduction
Every
human cell has a "second" genome, found in the cell's energy-generating
organelle, the mitochondrion. In fact, each mitochondrion has several
copies of its own genome, and there are several hundred to several thousand
mitochondria per cell. This means that the mitochondrial (mt) genome is
highly amplified. While each cell contains only two copies of a given
nuclear gene (one on each of the paired chromosomes), there are thousands
of copies of a given mt gene per cell. Because of this high copy number,
it is possible to obtain a mt DNA type from the equivalent of a single
cell's worth of mt DNA. Thus, mt DNA is the genetic system of choice in
cases where tissue samples are very old, very small, or badly degraded
by heat and humidity.
Under good circumstances - working from fresh cell samples - mt DNA is
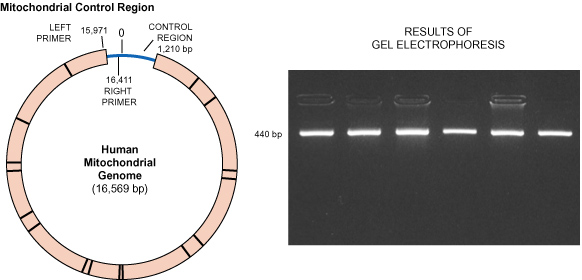
the easiest human DNA to amplify by PCR. This experiment examines a 440-nucleotide
sequence from the noncoding region of mt genome. Hand cycling is a realistic
alternative to automated thermal cyclers, and the high yield of amplified
product can be visualized in an agarose gel with a variety of stains.
Because each student is amplifying the same region, the gel electrophoresis
results will also be the same for each. However, amplified student samples
may be submitted to our Sequencing Service, which will generate student
mt DNA sequences and post the results on our Sequence Server. Comparison
of control region sequences reveals that most people have a unique pattern
of single nucleotide polymorphisms (SNPs). These sequence differences,
in turn, are the basis for far-ranging investigations on human DNA diversity
and the evolution of hominids.
Noncommercial, educational use only.